The Ebola epidemic could claim hundreds of thousands of lives and infect more than 1.4 million people by the end of January, according to a statistical forecast released this week by the U.S. Centers for Disease Control and Prevention.
The CDC forecast supports the drastically higher projections released earlier by a group of scientists, including epidemiologists with the Virginia Bioinformatics Institute, who modeled the Ebola spread as part of a National Institutes of Health-sponsored project called Midas, short for Models of Infectious Disease Agent Study.
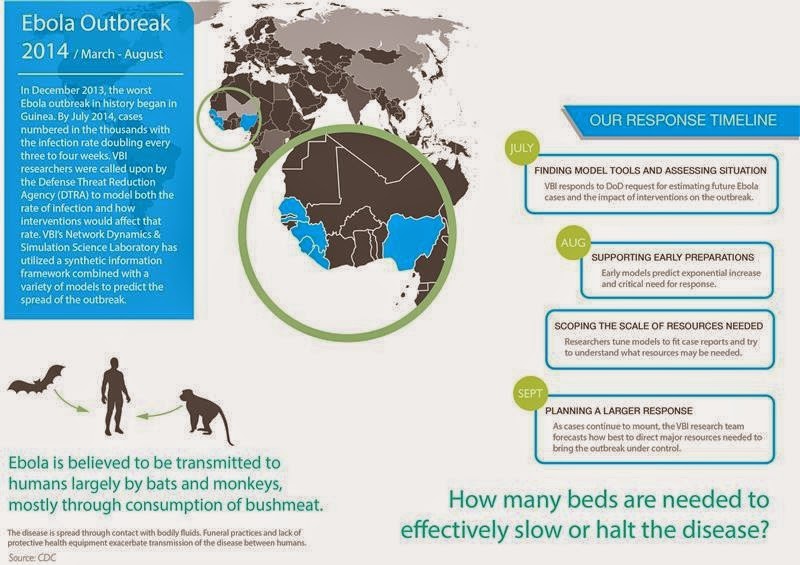
The effort is also supported by the federal Defense Threat Reduction Agency.
Before the scientists released results, the outbreak in West Africa was expected to be under control in nine months with only about 20,000 total cases. But modeling showed 20,000 people could be infected in just a single month.
The predictions could change dramatically if public health efforts become effective, but based on the virus's current uncontrolled spread, numbers of people infected could skyrocket.
"If the disease keeps spreading as it has been we estimate there could be hundreds of thousands of cases by the end of the year in Liberia alone," said Bryan Lewis, a computational epidemiologist with the Network Dynamics and Simulation Science Laboratory at the Virginia Bioinformatics Institute.
Lewis and his fellow researchers use a combination of models to predict outcomes of the epidemic.
The agent-based models are adaptive, evolving as more information is fed into them to provide an accurate forecast.
Pharmaceutical intervention, which is still on the horizon, is proving less effective in the models than supportive care and personal protection equipment for health care workers.
"The work with Ebola is not an isolated event," said Christopher Barrett, the executive director of the institute. "This research is part of a decades-long effort largely funded by the Defense Threat Reduction Agency to build a global synthetic population that will allow us to ask questions about our world and ourselves that we have never been able to ask before, and to use those answers to prevent or quickly intervene during a crisis."
Barrett and other institute leaders updated U.S. Sen. Tim Kaine and Virginia Tech President Timothy Sands about the Network Dynamics and Simulation Science Lab's role in analyzing the Ebola outbreak at the Virginia Tech Research Center in Arlington on Tuesday morning. That afternoon in Blacksburg they briefed staff members from U.S. Sen. Mark Warner's office.
A university-level Research Institute of Virginia Tech, the Virginia Bioinformatics Institute was established in 2000 with an emphasis on informatics of complex interacting systems scaling the microbiome to the entire globe. It helps solve challenges posed to human health, security, and sustainability. Headquartered at the Blacksburg campus, the institute occupies 154,600 square feet in research facilities, including state-of-the-art core laboratory and high-performance computing facilities, as well as research offices in the Virginia Tech Research Center in Arlington, Virginia.
Source: Virginia Tech